INRAE ECODIV Division
Deputy Chief (Chef de Département Adjoint)
UMR 1136 IAM
INRAE Centre Grand-Est, Nancy
Labex ARBRE
Bio: BSc in Biology at Bordeaux University (1995). MSc in Cell Biology and Physiology (1998) and PhD in Plant and Forest Sciences (2001) at Nancy University. Silver medal of the French Academy of Agriculture (2001). Hired research scientist in 2001 at INRA. 1 year sabbatical at Exeter University (2003-2004). HDR diploma from Lorraine University (2012). Region Lorraine Researcher Award (2012). Académie Stanislas Zivi Award (2012). INRA Research Director DR2 (2013). Appointed head of the Ecogenomic of interactions team in 2013, appointed Deputy Director (DUA) of the Tree-Microbe Interactions department in 2018. In charge of the WP1 (Integrative Biology) of the Lab of Excellence Labex ARBRE since 2018. Member of the SIReNa Doctoral School council at Université de Lorraine since 2020. Director (DU) of the INRAE/Université de Lorraine Tree–Microbe Interactions Joint Unit (UMR IAM) for the 2020-2023 period. Appointed Deputy Chief of the Ecology & Biodiversity Division (CDA Dept ECODIV) at INRAE in January 2024.
My early research was on functional genomics of the ectomycorrhizal symbiosis in the group of Francis Martin (INRA Nancy). I spent 1 year in Nick Talbot’s lab at Exeter University to extend my background in molecular phytopathology (2003-2004), and since 2005, my research in the Ecogenomics team has mostly focused on the poplar-poplar rust interaction. My research is part of a pluri-disciplinary scientific project called Mechanism and evolution of the poplar-poplar rust interaction which is transversal to the UMR IAM at INRAE Nancy.
Resume: My research at INRAE aims at elucidating how trees respond to pathogens and how biotroph pathogens are able to develop and feed on trees. Our model is the poplar-poplar rust pathosystem. I mainly use genomics and functional approaches to identify and characterize molecular determinants involved in disease resistance and defense response in poplar, and pathogenicity in the rust fungus Melampsora larici-populina as my main model pathosystem. In recent years, I have diversified my research in the frame of international projects using genomics in order to better understand the biology of rust fungi. Progress made in understanding rust fungal biotrophy will help setting new strategies to contain rust diseases.

Contact: sebastien.duplessis<at>inrae.fr / Phone, +33 383 39 40 13
Twitter: @SebbyRust
Links:
ResearchGate — Loop.Frontiers — Google Scholar
My research associates within the Tree/Microbe Interactions Department are Benjamin Petre, Nicolas Rouhier and Arnaud Hecker (IAM Redox and Stress Response team, Université de Lorraine); Fabien Halkett and Pascal Frey (IAM Forest Pathology team, INRAE); and Emmanuelle Morin (IAM Ecogenomics team, INRAE).
Current PhD and postdocs
• Emma Corre (PhD 2022-2025, co-supervisor Cécile Lorrain ETH Zurich). Impact of transposable elements invasion on genomes of Pucciniales (fungi causing rust diseases). Ecole Doctorale SIReNa.
• Ammar Abdalrahem (PhD 2022-2025, principal co-advisors P. Frey & F. Halkett). Evolutionary and functional studies of the variability of reproductive modes in the poplar rust fungus, Melampsora larici-populina. Ecole Doctorale SIReNa.
Former PhD and postdocs
• Cécile Rinaldi/Guinet (PhD, 2004-2007, co-advisor, F Martin), now Project Manager at ANSES, Laboratoire de Santé des Végétaux, Nancy, France
• Mehdi Kaytoue (PhD, 2007-2011, co-advisor, A Napoli, INRIA, LORIA Nancy), now Lecturer at LIRIS, Lyon, France
• Stéphane Hacquard (PhD, 2008-2011; post-doc fellow, 2012-2013, co-advisor, F Martin), now Group Leader at MPI, Koln, Germany
• Benjamin Petre (PhD CJS INRA 2009-2012, co-advisor, N Rouhier, post-doc CJS INRA 2013-2014 in S Kamoun’s lab at TSL, Norwich, UK); since 2018, lecturer at Lorraine University (UMR IAM)
• Natalya Saveleva (Post-doc ANR, 2013-2014). Employment unknown.
• Antoine Persoons (PhD, 2012-2015, co-advisor, S De Mita, F Halkett), post-doc in Diane Saunders at the JIC, Norwich, UK; then post-doc at UMR IAM Nancy.
• Cécile Lorrain (PhD CJS INRA 2014-2018, co-advisor A. Hecker); postdoc in Eva Stukenbrock Group in Plön/Kiel (Germany; 2018-2019, postdoc CJS INRA); postodc in Bruce McDonald‘s group, plant pathology, ETH Zurich; now Junior Project Leader, ETH Zurich.
• Si-Qi Tao (PhD at Beijing Forestry University, Advisor Prof. Ling-Mei Liang). 1-year stay at INRAE Nancy (2018-2019). International collaboration with Profs. Chengming Tian & Ling-Mei Liang. Now Lecturer at Beijing Forestry University.
• Clémentine Louet (PhD, 2018-2021, co-advisor, P. Frey). Caractérisation évolutive et fonctionnelle d’un locus d’avirulence chez l’agent de la rouille du peuplier, Melampsora larici-populina. Ecole Doctorale SIReNa. Now post-doc at UMR Bioger, EPLM team with Isabelle Fudal.
• Daniela Cardenas (visiting PhD student, University of Florida, 2021-2022; 6 months), supervisors Liliana Cano & Philippe Rott). Transcriptomics of the sugarcane-Puccinia kuehnii interaction.
• Claire Letanneur (PhD, 2017-2022). Co-advisor. International collaboration with Prof. Hugo Germain (Principal supervisor) at Université du Québec à Trois-Rivière. Host-specific effectors of the poplar rust fungus Melampsora larici-populina.
• Julie Lintz (PhD 2020-2023, co-advisor B. Petre). Caractérisation fonctionnelle et valorisation de peptides de défense multifonctionnels chez les arbres de la famille des Salicacées. Ecole Doctorale SIReNa. Now post-doc in S. Kamoun’s lab at TSL, Norwich, UK.

My current work focuses on…
– Populus and Melampsora genome analysis
– Rust comparative genomics (in the Melampsoraceae and Pucciniales, including the coffee rust Hemileia vastatrix and the Asian Soybean Rust Phakopsora pachyrhizi)
– Molecular basis of poplar-poplar rust R-Avr interactions
– Functional analysis of rust effectors in host cells
– Annotation and study of NB-LRR genes in Poplar and Oak genomes
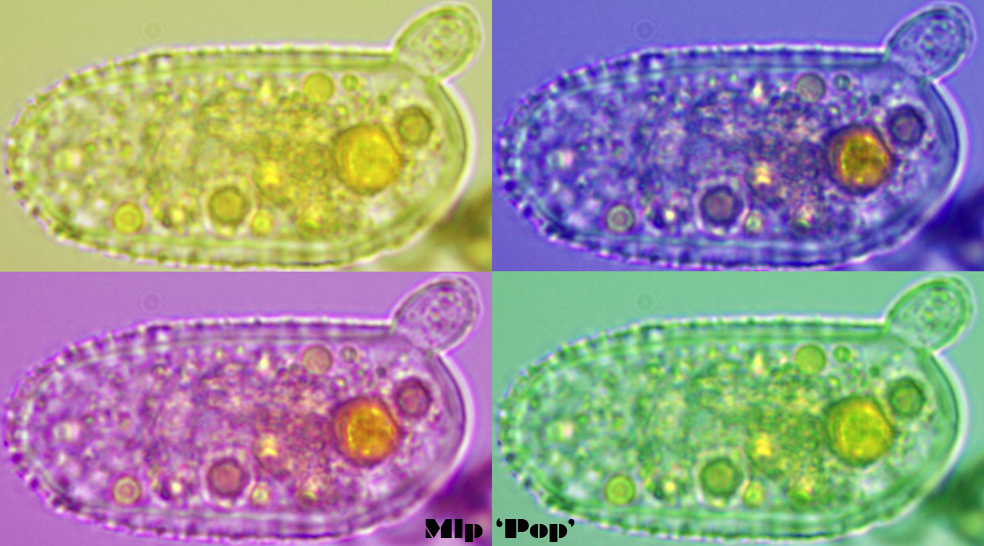
Melampsora larici-populina urediniospore germination
(© photo, Petre – Pop’art, Seb)
Active projects
• Rust genomics
Several collaborative projects are ongoing in the frame of the Community Science Program of the US DoE Joint Genome Institute (Walnut Creek, CA, USA):
> CSP2018 – Rust pangenomics: understanding the diversity and potential impact of rust fungi – a systematic collection of genomes & transcriptomes. International collaborative effort targeting 50 rust species across the Pucciniales order. Co-PIs are Sebastien Duplessis (INRA Nancy, France), M. Catherine Aime (Purdue Univ, USA), Melania Figueroa (Minnesota Univ, USA), Diane Saunders (JIC, UK), Guus Bakkeren (AAF, Canada), Les J. Szabo (USDA, USA), Robert F. Park (Sydney Univ, Australia), Ralf Vögele (Hohenheim Univ, Germany), Benjamin Schwessinger (ANU, Australia), Peter van Esse (TSL, UK), Peter N. Dodds (CSIRO, Australia).
> CSP2016 – Sequencing a reference genome for Phakopsora pachyrhizi, the fungal pathogen responsible for the Asian Soybean Rust. International collaborative effort. Co-PIs are Sebastien Duplessis (INRA Nancy, France), Catherine Sirven (Bayer Cropscience, Lyon, France), Francismar Correa Marcelino-Guimarães (EMBRAPA, Brazil), Ralf Voegele (Hohenheim Univ, Germany), Ulrich Schaffrath (Aachen Univ, Germany)
> CSP2014 – Combined population genomics and transcriptomics to decipher the molecular bases of virulence and host adaptation in the poplar leaf rust fungus Melampsora larici-populina. Co-PIs are Sebastien Duplessis (INRA Nancy, France), Pascal Frey (INRA Nancy, France), Fabien Halkett (INRA Nancy, France), Stephane De Mita (INRA Nancy, France).
In these projects we aim at understanding the biology of rust fungi and more particularly the complexity of their life cycle and their status of obligate biotroph through comparative genomics and transcriptomics including ab-initio and manual annotation of gene and transposable elements families. Most projects are conducted in the frame of international consortia. One of our current focus is comparative genomics of rust fungi in the Melampsoraceae family in collaboration with the JGI and Jana Sperschneider and Peter Dodds (CSIRO, Australia).
• Functional analysis of Candidate Effector Secreted Proteins (CSEPs).
Targeted CSEPs of Melamposra larici-populina are studied for their potential functions in the host cell. Heterologous systems such as the model plants Arabidopsis thaliana and Nicotiana benthamiana are used for localization and partner identification in the plant cell. Stable transformation of the poplar clone INRA 717-1-1B4 is used to validate localization and partners in poplar. Recombinant CSEPs are produced in E. coli and their structures are studied by NMR. Locally, Arnaud Hecker and Nicolas Rouhier (Lorraine Univ, France) are key actors and this project involves past and ongoing collaborations with Sophien Kamoun (TSL, UK), Hugo Germain (Trois-Rivières Univ, Quebec, Canada), Andre Padilla and Karine de Guillen (CNRS/INSERM, Montpellier, France).
• Tree genomics – Annotation of NB-LRR gene families in model tree species.
NB-LRR gene families have been identified and annotated in the genomes of the tree species Populus trichocarpa and Quercus robur. The gene catalog of poplar NB-LRR genes has been completed and revised through the different genome versions and we aim to provide an update when the poplar genome assembly v4 will be provided to the community. We also aim to use the RenSeq approach developed at the TSL (Norwich, UK) to complete our estimate of this gene family in poplar. The gene catalog of oak NB-LRR genes is being scrutinized in the frame of the ANR GENOAK project (Coord. Christophe Plomion, INRAE, France).
• Tree-rust interaction – Study of defense peptides in poplar.
A rust-induced secreted peptide (RISP) encoding gene was identified in poplar upon infection by the poplar rust fungus Melampsora larici-populina. RISP displays dual activities with antifungal and immunomodulatory properties. Our current work focuses on these properties (project coordinated by Benjamin Petre).
Publications
2024
• LINTZ J, GOTO Y, BENDER K, BCHINI R, DUBRULLE G, CAWSTON E, ZIPFEL C, DUPLESSIS S, PETRE B. Genetically-clustered antifungal phytocytokines and receptor proteins function together to trigger plant immune signaling. Submitted in 2023. Available as a BioRxiv: doi: https://doi.org/10.1101/2023.11.27.568785.
• LETANNEUR C, BRISSON A, BISAILLON M, DEVEZE T, PLOURDE MB, SCHATTAT M, DUPLESSIS S, GERMAIN H (2024) Host-specific and homologous pairs of Melampsora larici-populina effectors unveil novel Nicotiana benthamiana stromule induction factors. Molecular Plant-Microbe Interactions. 37: 277-289. doi: 10.1094/MPMI-09-23-0148-FI.
2023
• LOUET C, SAUBIN M, ANDRIEUX A, PERSOONS A, GORSE M, PETROWSKI J, FABRE B, DE MITA S, DUPLESSIS S, FREY P, HALKETT F (2023) A point mutation and a large deletion at the candidate avirulence locus AvrMlp7 in the poplar rust fungus correlate with the poplar RMlp7 resistance breakdown. Molecular Ecology. 32:2472–2483. Special issue Evolutionary ecology of human-associated microbes. doi: https://doi.org/10.1111/mec.16294.
• GUPTA YK, MARCELINO-GUIMARAES FC, LORRAIN C, FARMER A, HARIDAS S, FERREIRA EGC, LOPES-CAITAR VS, SANTANA OLIVEIRA L, MORIN E, WIDDISON S, CAMERON C, INOUE Y, THOR K, ROBINSON K, DRULA E, HENRISSAT B, LABUTTI K, RUDSIT BINI AM, PAGET E, SINGAN V, DAUM C, DORME C, VAN HOEK M, JANSSEN A, CHANDAT L, TARRIOTTE Y, RICHARDSON J, ARAUJO MELO BDV, WITTENBERG AHJ, SCHNEIDERS H, PEYRARD S, GOULART ZANARDO L, HOLTMAN VC, COULOMBIER-CHAUVEL F, LINK TI, BALMER D, MULLER AN, KIND S, BOHNERT S, WIRTZ L, CHEN C, YAN M, NG V, GAUTIER P, MEYER MC, VOEGELE RT, LIU Q, GRIGORIEV IV, CONRATH U, BROMMONSCHENKEL SH, LOEHRER M, SCHAFFRATH U, SIRVEN C, SCALLIET G*, DUPLESSIS S*, VAN ESSE HP* (2023) Major proliferation of transposable elements shaped the genome of the soybean rust pathogen Phakopsora pachyrhizi. Nature communications 14: 1835. *Co-Corresponding author. doi: 10.1038/s41467-023-37551-4. Available as a BioRxiv.
• GUERILLOT P, SALAMOV A, LOUET C, MORIN E, FREY P, GRIGORIEV IV, DUPLESSIS S* (2023) A remarkable expansion of oligopeptide transporter genes in rust fungi (Pucciniales) suggests a specialization in nutrients acquisition for obligate biotrophy. Phytopathology 113: 252-264. doi: 10.1094/PHYTO-04-22-0128-R. Available as a BioRxiv: (10.1101/2022.04.20.488971v1). *Corresponding author.
2022
• DUPLESSIS S (2022) Exploring genomic determinants of host specialization in Botrytis cinerea. Recommendation for article ‘Botrytis cinerea strains infecting grapevine and tomato display contrasted repertoires of accessory chromosomes, transposons and small RNAs’ by Simon et al. doi:10.24072/pci.genomics.100023. Peer Community in Genomics, 100023.
• LOUET C, DUPLESSIS S, FREY P, PETRE B (2022) A survey of highly influential studies on plant pathogen effectors during the last two decades (2000-2020). Frontiers in Plant Science 13: 920281. doi: 10.3389/fpls.2022.920281. Available as a HAL archive (hal-03386375). Part of the research topic Secretomics: More Secrets to Unravel on Plant-Fungus Interactions, Volume II.
• PETRE B*, DUPLESSIS S* (2022) A decade after the first Pucciniales genomes : a bibliometric snapshot of (post) genomics studies in three model rust fungi.Frontiers in Microbiology 13: 989580. doi: 10.3389/fmicb.2022.989580. Mini-review in Research Topic Celebrating 20 Years of Microbial Plant Pathogen Genomes. Available as a HAL archive hal-03719656. *Co-corresponding author.
• LINTZ J, DUBRULLE G, CAWSTON E, DUPLESSIS S, PETRE B (2022) A short review of anti-rust fungi peptides: diversity, bioassays, and unknown mode of actions. Frontiers in Agronomy 4: 966211. doi: 10.3389/fagro.2022.966211. Mini-review in Research Topic Advances in Novel Natural Product Pesticides.
• PETRE B, LOUET C, LINTZ J, DUPLESSIS S (2022) 2000-2019: twenty years of highly influential publications in plant immunity. Molecular Plant-Microbe Interactions 9: 748-754. doi: 10.1094/MPMI-05-22-0112-CR. Available as a HAL archive (hal-03325099v2).
• LOUET C, BLOT C, SHELEST E, GUERILLOT P, SALAMOV A, ZANNINI F, PETROWSKI J, GRIGORIEV IV, FREY P, DUPLESSIS S* (2022) Annotation survey and life cycle transcriptomics of transcription factors in rust fungi (Pucciniales) identify a possible role for cold shock proteins in dormancy exit. Fungal Genetics & Biology 1061: 103698. Available as a BioRxiv (10.1101/2021.10.19.465044v1). *Corresponding author.
• TAO S, ZHANG Y, QU L, TIAN C, DUPLESSIS S, ZHANG N (2022) Elevated ozone concentration and nitrogen addition increase poplar rust severity by shifting the phyllosphere microbial community. Journal of Fungi 8: 523. doi: 10.3390/jof8050523. Available as a BioRxiv (10.1101/2021.12.23.474070v1)
• DUPLESSIS S (2022) Comparative genomics in the chestnut blight fungus Cryphonectria parasitica reveals large chromosomal rearrangements and a stable genome organization. Recommendation for article ‘Chromosomal rearrangements with stable repertoires of genes and transposable elements in an invasive forest-pathogenic fungus’ by Demené et al. doi:10.24072/pci.genomics.100013. Peer Community in Genomics, 100013.
• PERSOONS A, MAUPETIT A, LOUET C, ANDRIEUX A, LIPZEN A, BARRY KW, NA H, ADAM C, GRIGORIEV I, SEGURA V, DUPLESSIS S, FREY P, HALKETT F, DE MITA S (2022). Genomic signatures of a major adaptive event in the pathogenic fungus Melampsora larici-populina. Genome Biology and Evolution. 14: evab279. doi: 10.1093/gbe/evab279. Available as a BioRxiv, doi: 10.1101/2021.04.09.439223v1.
2021
• GUERILLOT P, MORIN E, KOHLER A, LOUET C, DUPLESSIS S* (2021) A comprehensive collection of transcriptome data to support life cycle analysis of the poplar rust fungus Melampsora larici-populina. Brief research data report. 4p. Available as a BioRxiv. *Corresponding author. doi: 10.1101/2021.07.25.453710. Related datasets available at FigShare doi: 10.6084/m9.figshare.15048822.v1.
• WANG X, ZHUANG R, ZHAI T, ZHANG X, TANG C, ZHAO H, CHENG Y, XU Q, WANG J, DUPLESSIS S, KANG Z, WANG X (2021) Two stripe rust effectors suppress wheat resistance by preventing import of a host Fe-S protein into chloroplasts. Plant Physiology. 187: 2530-2543.
• DUPLESSIS S, LORRAIN C, PETRE B, FIGUEROA M, DODDS PN, AIME MC (2021) Host adaptation and virulence in heteroecious rust fungi. Annual Review of Phytopathology. 59: 403-422. Corresponding author. doi: 10.1146/annurev-phyto-020620-121149
• LORRAIN C, OGGENFUSS U, CROLL D, DUPLESSIS S, STUKENBROCK EH (2021) Transposable elements in fungi: coevolution with the host genome shapes genome architecture, plasticity and adaptation. In: Encyclopedia of Mycology Vol.1: 142-155. 10.1016/B978-0-12-819990-9.00042-1
2020
• PETRE B, LORRAIN C, STUKENBROCK EH, DUPLESSIS S (2020) Host-specialized transcriptome of plant associated organisms. Current Opinion in Plant Biology. 56: 81-88. Corresponding author. doi: 10.1016/j.pbi.2020.04.007
• TAO SQ, AUER L, MORIN E, LIANG Y-M, DUPLESSIS S (2020) Transcriptome analysis of apple leaves infected by the rust fungus Gymnosporangium yamadae at two sporulation stages. Molecular Plant-Microbe Interactions doi: 10.1094/MPMI-07-19-0208-R. Co-corresponding author. Available as a bioRxiv, doi: https://doi.org/10.1101/717058.
2019
• DEGUILLEN K, LORRAIN C, TSAN P, BARTHE P, PETRE B, SAVELEVA N, ROUHIER N, DUPLESSIS S, PADILLA A, HECKER A (2019) Structural genomic applied on the rust fungus Melampsora larici-populina reveals two candidate effector proteins adopting cystine-knot and nuclear transport factor 2-like protein folds. Scientific reports 9:18084. Available as a bioRxiv, doi: https://doi.org/10.1101/727933.
• TAO SQ, CAO B, MORIN E, LIANG Y-M, DUPLESSIS S (2019) Comparative transcriptomics of Gymnosporangium spp. teliospores reveals a conserved genetic program at this specific stage of the rust fungal life cycle. BMC Genomics 20:723. Co-corresponding author.
• LORRAIN C, GONCALVES DOS SANTOS KC, GERMAIN H, HECKER A, DUPLESSIS S (2019) Advances in understanding obligate biotrophy in rust fungi. Tansley review, New Phytologist. doi: 10.1111/nph.15641. Corresponding author.
2018
• AHMED B, GONCALVES DOS SANTOS KC, SANCHEZ IB, PETRE B, LORRAIN C, DUPLESSIS S, DESGAGNE-Penix I, GERMAIN H (2018) A rust fungal effector binds plant DNA and modulates transcription. Scientific reports, 8: 14718.
• PLOMION C, AURY JM, AMSELEM J, LEROY T, MURAT F, DUPLESSIS S, FAYE S, FRANCILLONNE N, LABADIE K,LE PROVOST G, LESUR I, BARTHOLOMÉ J, FAIVRE-RAMPANT P, KOHLER A, LEPLÉ J-C, CHANTRET N, CHEN J, DIÉVART A, ALAEITABAR I, BARBE V, BELSER C, BERGÈS H, BODÉNÈS C, BOGEAT-TRIBOULOT MB, BOUFFAUD M-L, BRACHI B, COHEN D, COULOUX A, DA SILVA C, DOSSAT C, EHRENMANN F, GASPIN C, GRIMA-PETTENATI J, GUICHOUX E HECKER A, HERRMANN S, HUGUENEY P, HUMMEL I, KLOPP C, LALANNE C, LASCOUX M, LASSERRE E, LEMAINQUE A, DESPREZ-LOUSTAU M-L, LUYTEN I, MADOUI MA, MANGENOT S, MARCHAL C, MAUMUS F, MERCIER J, MICHOTEY C, PANAUD O, PICAULT N, ROUHIER N, RUÉ O, RUSTENHOLZ C, SALIN F, SOLER M, TARKKA M, VELT A, ZANNE A, MARTIN F, WINCKER P, QUESNEVILLE H, KREMER A, SALSE J (2018) Oak genome reveals facets of long lifespan. Nature Plants, doi: 10.1038/s41477-018-0172-3.
• ZENG Z, SUN H, VAINIO AJ, RAFFAELLO T, KOVALCHUK A, MORIN E, DUPLESSIS S, ASIEGBU FO (2018) Intraspecific comparative genomics of isolates of the Norway spruce pathogen (Heterobasidion parviporum) and identification of its potential virulence factors. Research article, BMC Genomics, 19: 220.
• LORRAIN C, PETRE B, DUPLESSIS S (2018) Show me the way: rust effector targets in heterologous plant systems. Current Opinion in Microbiology, 46:19-25. Corresponding author.
• LORRAIN C, MARCHAL C, HACQUARD S, DELARUELLE C, PETROWSKI J, PETRE B, HECKER A, FREY P, DUPLESSIS S (2018) The rust fungus Melampsora larici-populina expresses a conserved genetic program and distinct sets of secreted protein genes during infection of its two host plants, larch and poplar. Molecular Plant-Microbe Interactions. doi: 10.1094/MPMI-12-17-0319-R. July issue cover. Corresponding author.
Early version available at BioRxiv.
• GERMAIN H, JOLY DL, MIREAULT C, PLOURDE MB, LETANNEUR C, STEWART D, MORENCY M-J, PETRE B, DUPLESSIS S, SEGUIN A (2018) Infection assays in Arabidopsis reveal candidate effectors from the poplar rust fungus that promote susceptibility to bacteria and oomycete pathogens. Molecular Plant Pathology. 19: 191-200. doi: 10.1111/mpp.12514.
2017
• AIME MC, McTAGGART AR, MONDO SJ, DUPLESSIS S (2017) Phylogenetics and phylogenomics of rust fungi. In: Fungal phylogenetics and phylogenomics. Townsend JP & Wang Z, Eds. Elsevier Academic Press, Cambridge, MA, USA. Advances in Genetics Vol 100: 267-307. Corresponding author.
• SPERSCHNEIDER J, TAYLOR J, DODDS PN, DUPLESSIS S (2017) Computational methods for predicting effectors in rust pathogens. Wheat Rust Disease – Methods and Protocols. Methods in Molecular Biology. Springer, 2017, pp73-83.
2016
• DUPLESSIS S, BAKKEREN G, JOLY, DL, eds. (2016) Genomics Research on Non-model Plant Pathogens: Delivering Novel Insights into Rust Fungus Biology. Lausanne: Frontiers Media. doi: 10.3389/978-2-88919-814-6. Collective Ebook. Co-editor.
• BAKKEREN G, JOLY DL, DUPLESSIS S (2016) Editorial: Genomics Research on Non-model Plant Pathogens: Delivering Novel Insights into Rust Fungus Biology. Frontiers in Plant Science 7:216. doi: 10.3389/fpls.2016.00216. Co-corresponding author.
• PETRE B, SAUNDERS DGO, SKLENAR J, LORRAIN C, KRASILEVA KV, WIN J, DUPLESSIS S, KAMOUN S (2016) In planta expression screens of candidate effector proteins from the wheat yellow rust fungus reveal processing bodies as a pathogen-targeted plant cell compartment. PLoS ONE, 11(2):e0149035. Pre-print @ bioRxiv.
• PETRE B, HECKER A, GERMAIN H, TSAN P, SKLENAR J, PELLETIER G, SEGUIN A, DUPLESSIS S, ROUHIER N (2016) The poplar rust-induced secreted protein (RISP) inhibits the growth of the leaf rust pathogen Melampsora larici-populina and triggers cell culture alkalinisation. Frontiers in Plant Science 7:97 doi: 10.3389/fpls.2016.00097. Co-Corresponding author.
• PLOMION C, BASTIEN C, BOGEAT-TRIBOULOT M-B, BOUFFIER L, DEJARDIN A, DUPLESSIS S, FADY B, HEUERTZ M, LE GAC A-L, LE PROVOST G, LEGUE V, LELU-WALTER M-A, LEPLE J-C, MAURY S, MOREL A, ODDOU-MURATORIO S, PILATE G, SANCHEZ L, SCOTTI I, SCOTTI-SAINTAGNE C, SEGURA V, TRONTIN J-F, VACHER C (2016) Forest tree genomics: 10 achievements from the past 10 years and future prospects. Annals of Forest Science, 73:77-103 doi: 10.1007/s13595-015-0488-3.
• PLOMION C, AURY J-M, AMSELEM J, ALAEITABAR T, BARBE V, BELSER C, BERGES H, BODENES-BREZARD C, BOUDET N, BOURY C, CANAGUIER A, COULOUX A, DA SILVA C, DUPLESSIS S, EHRENMANN F, ESTRADA-MAIREY B, FOUTEAU S, FRANCILLONNE N, GASPIN C, GUICHARD C, KLOPP C, LABADIE K, LALANNE C, LE CLAINCHE I, LEPLÉ JC, LE PROVOST G, LEROY T, LESUR-KUPIN I, MARTIN F, MERCIER J, MICHOTEY C, MURAT F, SALIN F, STEINBACH D, FAIVRE-RAMPANT P, WINCKER P, SALSE J, QUESNEVILLE H, KREMER A (2016) Decoding the oak genome: public release of sequence data, assembly, annotation and publication strategies. Molecular Ecology Resources 16:254-265.
2015
• LORRAIN C, HECKER A, DUPLESSIS S (2015) Effector-mining in the poplar rust fungus Melampsora larici-populina secretome. Mini-review, Frontiers in Plant Science 6:1051. Corresponding author.
• SILVA DN, DUPLESSIS S, TALHINHAS P, AZINHEIRA H, PAULO OS, BATISTA D (2015) Genomic Patterns of Positive Selection at the Origin of Rust Fungi. PLoS One 10:e0143959.
• PETRE B, LORRAIN C, SAUNDERS DG, WIN J, SKLENAR J, DUPLESSIS S, KAMOUN S (2015) Rust fungal effectors mimic host transit peptides to translocate into chloroplasts. Cellular Microbiology 18:453-465 doi: 10.1111/cmi.12530.
• PERLIN MH, AMSELEM J, FONTANILLAS E, TOH SS, CHEN Z, GOLDBERG J, DUPLESSIS S, HENRISSAT B, YOUNG S, ZENG Q, AGUILETA G, PETIT E, BADOUIN H, ANDREWS J, RAZEEQ D, GABALDÓN T, QUESNEVILLE H, GIRAUD T, HOOD ME, SCHULTZ DJ, CUOMO CA (2015) Sex and parasites: genomic and transcriptomic analysis of Microbotryum lychnidis-dioicae, the biotrophic and plant-castrating anther smut fungus. BMC Genomics 16: 461.
• PETRE B, SAUNDERS DGO, SKLENAR J, LORRAIN C, WIN J, DUPLESSIS S, KAMOUN S (2015) Candidate effector proteins of the rust pathogen Melampsora larici-populina target diverse plant cell compartments. Molecular Plant-Microbe Interactions. 28:689-700.
• LORRAIN C, MORIN E, PERSOONS A, HECKER A, DUPLESSIS S (2015) Les effecteurs de Melampsora larici-populina. Actes du séminaire 2015 de l’Ecole Doctorale RP2E, Nancy, France (ISBN 2-9518564-8-2), 8 pages.
2014
• DUPLESSIS S, BAKKEREN G, HAMELIN R (2014) Advancing knowledge on biology of rust fungi through genomics. Advances in Botanical Research 70:173-209.
• PEGEOT H, KOH CS, PETRE B, MATHIOT S, DUPLESSIS S, HECKER A, DIDIERJEAN C, ROUHIER N (2014) The poplar Phi class glutathione transferase: expression, activity and structure of GSTF1. Frontiers in Plant Science 5:712.
• PERNACI M, DE MITA S, ANDRIEUX A, PETROWSKI J, HALKETT F, DUPLESSIS S, FREY P (2014) Genome-wide patterns of segregation and linkage disequilibrium: the construction of a linkage genetic map of the poplar rust fungus Melampsora larici-populina. Frontiers in Plant Science 5:454.
• PERSOONS A, MORIN E, DELARUELLE C, PAYEN T, HALKETT F, FREY P, DE MITA S, DUPLESSIS S (2014) Patterns of genomic variation in the poplar rust fungus Melampsora larici-populina identify pathogenesis-related factors. Frontiers in Plant Science, 5:450. Corresponding author.
• PETRE B, JOLY DL, DUPLESSIS S (2014) Effector proteins of rust fungi. Mini-review, Frontiers in Plant Science 5: 416. Corresponding author.
• LOEHRER M, VOGEL A, HUETTEL B, REINHARDT R, BENES V, DUPLESSIS S, USADEL, B, SCHAFFRATH U (2014) On the current status of Phakopsora pachyrhizi genome sequencing. Perspective, Frontiers in Plant Science 5:377.
• PETRE B, HACQUARD S, DUPLESSIS S, ROUHIER N (2014) Poplar Leucine-Rich Repeat Receptor-Like Protein (LRR-RLP) genes cluster with genes coding putative endogenous ligands. Frontiers in Plant Science 5:111. Co-Corresponding author.
• TALHINAS P, AZINHEIRA HG, VIEIRA B, LOUREIRO A, TAVARES S, BATISTA D, MORIN E, PETITOT A-S, PAULO OS, POULAIN J, DA SILVA C, DUPLESSIS S, SILVA MdC, FERNANDEZ D (2014) Overview of the functional virulent genome of the coffee leaf rust pathogen Hemileia vastatrix with an emphasis on early stages of infection. Frontiers in Plant Science 5:88.
2013
• HACQUARD S, DELARUELLE C, FREY P, TISSERANT E, KOHLER A, DUPLESSIS S (2013) Transcriptome analysis of poplar rust telia reveals overwintering adaptation and tightly coordinated karyogamy and meiosis processes. Frontiers in Plant Science 4: 456. Corresponding author.
• DUPLESSIS S, SPANU PD, SCHIRAWSKI J (2013) Biotrophic fungi (powdery mildews, Rusts and Smuts). In Ecological genomics of the fungi, Plant-interacting fungi section. Martin F (ed), Wiley-Blackwell. 149-168.
• FERNANDEZ D, TALHINHAS P, DUPLESSIS S (2013) Rust fungi achievements and future challenges on genomics and host-parasite interactions. The Mycota vol. XI, Agricultural applications 2nd edition, Kempken F ed, Springer. 315-342.
• PERSOONS A, HALKETT F, DUPLESSIS S, DE MITA S (2013) La génomique des populations : évolution des méthodes de détection de la sélection. Actes du séminaire 2013 de l’Ecole Doctorale RP2E, Nancy, France (ISBN 2-9518564-8-2). 8 pages.
2012
• PETRE B, MORIN E, TISSERANT E, HACQUARD S, DA SILVA C, POULAIN J, DELARUELLE C, ROUHIER N, MARTIN F, KOHLER A, DUPLESSIS S (2012) RNA-Seq of early-infected poplar leaves by the rust pathogen Melampsora larici-populina uncovers PtSultr3;5, a fungal-induced host sulfate transporter. PLoS ONE 7(8): e44408. Corresponding author.
• HAMEL L-P, NICOLE M-C, DUPLESSIS S, ELLIS BE (2012) MAPK signaling in plant-interacting fungi: distinct messages from conserved messengers. The Plant Cell, 24:1327-1351.
• OLSON A, AERTS A, ASIEGBU F, BELBAHRI L, BOUZID O, BROBERG A, CANBACK B, COUTINHO PM, CULLEN D, DALMAN K, DEFLORIO G, VAN DIEPEN LTA, DUNAND C, DUPLESSIS S, DURLING M, GONTHIER P, GRIMWOOD J, GUNNAR FOSSDAL C, HANSSON D, HENRISSAT B, HIETALA A, HIMMELSTRAND K, HOFFMEISTER D, HOGBERG N, JAMES TY, KARLSSON M, KOHLER A, KUES U, LEE Y-H, LIN Y-C, LIND M, LINDQUIST E, LOMBARD V, LUCAS S, LUNDEN K, MORIN E, MURAT C, PARK J, RAFFAELLO T, ROUZE P, SALAMOV A, SCHMUTZ J, SOLHEIM H, STAHLBERG J, VELEZ H, DE VRIES RP, WIEBENGA A, WOODWARD S, YAKOVLEV I, GARBELOTTO M, MARTIN F, GRIGORIEV IV, STENLID J (2012) Insight into trade-off between wood decay and parasitism from the genome of a fungal forest pathogen. The New Phytologist, 194:1001-1013.
 • HACQUARD S, JOLY DJ, LIN Y-C, TISSERANT E, FEAU N, DELARUELLE C, LEGUE V, KOHLER A, TANGUAY P, PETRE B, FREY P, VAN DE PEER Y, ROUZE P, MARTIN F, HAMELIN RC, DUPLESSIS S (2012)A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici-populina (poplar leaf rust). Molecular Plant-Microbe Interactions, 25:279-293. Corresponding author.
• HACQUARD S, JOLY DJ, LIN Y-C, TISSERANT E, FEAU N, DELARUELLE C, LEGUE V, KOHLER A, TANGUAY P, PETRE B, FREY P, VAN DE PEER Y, ROUZE P, MARTIN F, HAMELIN RC, DUPLESSIS S (2012)A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici-populina (poplar leaf rust). Molecular Plant-Microbe Interactions, 25:279-293. Corresponding author. • FERNANDEZ D, TISSERANT E, TALHINHAS P, AZINHEIRA H, VIEIRA A, PETITOT A-S, LOUREIRO A, SILVA MdC, POULAIN J, DA SILVA C, DUPLESSIS S (2012) 454-pyrosequencing of Coffea arabica leaves infected by the rust fungus Hemileia vastatrix reveals in planta expressed pathogen-secreted proteins and plant functions expressed in a late compatible plant-rust interaction. Molecular Plant Pathology, 13:17-37. Corresponding author.
• SILVA DN, VIEIRA A, TALHINHAS P, AZINHEIRA HG, SILVA MDC, FERNANDEZ D, DUPLESSIS S, PAULO OS, BATISTA D (2012) Phylogenetic analysis of Hemileia vastatrix and related taxa using a genome-scale approach. In Proccedings of the 24th Conference of the Association for Science and Information on Coffee. In press.
• DOWKIW A, JORGE V, VILLAR M, VOISIN E, GUERIN V, FAIVRE-RAMPANT P, BRESSON A, BITTON F, DUPLESSIS S, FREY P, PETRE B, GUINET C, XHAARD C, FABRE B, HALKETT F, PLOMION C, LALANNE C, BASTIEN C (2012) Breeding poplars with durable resistance to Melampsora larici-populina leaf rust: a multidisciplinary approach to understand and delay pathogen adaptation. Extended abstract in the Proceedings of the 4th International Workshop on Genetics of Host-Parasite Interactions in Forestry, Oregon, USA, General technical report PSW-GTR-240 Forest Service USDA, Eds. Pp-31-38.
2011
• HACQUARD S, PETRE B, FREY P, HECKER A, ROUHIER N, DUPLESSIS S (2011) The poplar-poplar rust interaction: insights from genomics and transcriptomics. Journal of Pathogens. (edited by B Tyler). Vol. 2011, Article ID 716041, 11 pages (doi:10.4061/2011/716041). Corresponding author.
• L’HARIDON F, AIME S, DUPLESSIS S, ALABOUVETTE C, STEINBERG C, OLIVAIN C (2011) Isolation of differentially expressed genes during interactions between tomato cells and a protective or a non-protective strain of Fusarium oxysporum. Physiological and Molecular Plant Pathology, 76:9-19.
• VIEIRA A, TALHINHAS P, LOUREIRO A, DUPLESSIS S, FERNANDEZ D, SILVA MdC, PAULO OS, AZINHEIRA HG (2011) Validation of real time quantitative PCR reference genes for in planta expression studies in Hemileia vastatrix, the causal agent of coffee leaf rust. Fungal Biology, 115:891-901.
• DUPLESSIS S, CUOMO CA, LIN Y-C, AERTS A, TISSERANT E, VENEAULT-FOURREY C, JOLY DL, HACQUARD S, AMSELEM J, CANTAREL B, CHIU R, COUTINHO P, FEAU N, FIELD M, FREY P, GELHAYE E, GOLDBERG J, GRABHERR M, KODIRA C, KOHLER A, KUES U, LINDQUIST EA, LUCAS S, MAGO R, MAUCELI E, MORIN E, MURAT C, PANGILINAN JL, PARK R, PEARSON M, QUESNEVILLE H, ROUHIER N, SAKTHIKUMAR S, SCHMUTZ J, SELLES B, SHAPIRO H, TANGUAY P, TUSKAN GA, HENRISSAT B, VAN DE PEER Y, ROUZE P, ELLIS JG, DODDS PN, SCHEIN JE, ZHONG S, HAMELIN RC, GRIGORIEV IV, SZABO LJ, MARTIN F (2011) Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proceedings of the National Academy of Science USA, 108:9166-9171. Open Access. Co-1st author. Co-corresponding author.
• PETRE B, MAJOR I, ROUHIER N, DUPLESSIS S (2011)Genome-wide analysis of eukaryote thaumatin-like proteins (TLPs) with an emphasis on poplar. BMC Plant Biology, 11:33. Highly accessed. Corresponding author.
• KAYTOUE M, DUPLESSIS S, NAPOLI A (2011) Toward the discovery of itemsets with significant variations in gene expression matrices. In B Fichet et al eds, Classification and Multivariate Analysis for Complex Data Structures, Studies in Classification, Data Analysis, and Knowledge Organisation (Springer). Pp-465-473. DOI: 10.1007/978-3-642-13312-1_49
• PLOMION C, VINCENT D, BEDON F, JOETS J, BONHOMME J, MORABITO D, DUPLESSIS S, NILSSON R, WINGSLE G, LARSSON C, JOLIVET Y, RENAUT J, PECHANOVA O, YUCEER C (2011) Poplar proteomics: update and future challenges. In Genetics, genomics and breeding of Poplar, Joshi CP, DiFazio SP & Kole C (eds), Science Publishers, Enfield, New Hampshire, USA. Pp-128-165.
• TALHINHAS P, AZINHEIRA HG, LOUREIRO A, BATISTA D, VIEIRA B, PINA-MARTINS F, TISSERANT E, PETITOT A-S, PAULO OS, DUPLESSIS S, SILVA MC, FERNANDEZ D (2011) Overview of the functional virulent genome of the coffee leaf rust pathogen Hemileia vastatrix. In Proccedings of the 23rd Conference of the Association for Science and Information on Coffee. pp-414-422.
• PETRE B, MAJOR I, FREY P, MARTIN F, ROUHIER N, DUPLESSIS S (2011) Les apports de la génomique pour l’étude des familles multigéniques: exemple des thaumatin-like proteins (TLPs) chez le peuplier. Actes du séminaire 2011 de l’Ecole Doctorale RP2E, Nancy, France (ISBN 2-9518564-8-2). 8 pages.
2010
• HACQUARD S, DELARUELLE C, LEGUE V, TISSERANT E, KOHLER A, FREY P, MARTIN F, DUPLESSIS S (2010) Laser capture microdissection of uredinia formed by Melampsora larici-populina revealed a transcriptional switch between biotrophy and sporulation. Molecular Plant-Microbe Interactions23:1275-1286. Corresponding author.
• MARTIN F, KOHLER A, MURAT C, BALESTRINI R, COUTINHO PM, JAILLONO, MONTANINIB, MORINE, NOEL B, PERCUDANI R, PORCEL B, RUBINIA, AMICUCCI A, AMSELEMJ, ANTHOUARDV, ARCIONIS, ARTIGUENAVEF, AURYJM, BALLARIOP, BOLCHIA, BRENNAA, BRUNA, BUEEM, CANTARELB, CHEVALIERG, COULOUXA, DA SILVAC, DENOEUDF, DUPLESSISS, GHIGNONE S, HILSELBERGERB, IOTTIM, MARÇAISB, MELLOA, MIRANDAM, PACIONIG, QUESNEVILLEH, RICCIONIC, RUOTOLOR, SPLIVALLOR, STOCCHI V, TISSERANTE, VISCOMIAR, ZAMBONELLIA, ZAMPIERIE, HENRISSATB, LEBRUNMH, PAOLOCCIF, BONFANTEP, OTTONELLO S, WINCKERP (2010) Périgord Black Truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 464:1033-1038.
• DUPLESSIS S and colleagues. (2010) Abstracts of oral and poster presentations given at the 4th International Rusts of Forest Trees Conference, Florence, Italy, 3-6 May 2010 (Contributor to 3 presentations and 2 posters). Phytopathologia Mediterranea 49:421-434.
• JOLY DL, HACQUARD S, FEAU N, TANGUAY P, MARTIN F, HAMELIN R, DUPLESSIS S (2010) Chasing effectors in the secretome of Melampsora larici-populina. Paper 77 In Biology of plant-microbe interactions, Vol. 7, H. Antoun, T. Avis, L. Brisson, D. Prévost, and M. Trepanier, eds. International Society for Molecular Plant-Microbe Interactions, St. Paul, MN. Proceedings of the 2009 IS-MPMI congress in Québec. Corresponding author.
2009
• GUERRA F, DUPLESSIS S, KOHLER A, MARTIN F, TAPIA J, LEBED P, ZAMUDIO F, GONZALEZ E (2009) Gene expression analysis of Populus deltoides roots subjected to copper stress. Environmental and Experimental Botany 67:335-344.
• DUPLESSIS S, MAJOR I, MARTIN F, SEGUIN A (2009) Poplar and pathogen interactions: insights from genome-wide analyses of resistance and defense gene families and expression profiling of poplar leaves upon infection with rust. Critical Reviews in Plant Science 28:309-334. Review. Corresponding author.
• KEMPPAINEN M, DUPLESSIS S, MARTIN F, PARDO AG (2009) RNA silencing in the model mycorrhizal fungus Laccaria bicolor: gene Knock-down of nitrate reductase results in inhibition of symbiosis with Populus. Environnemental Microbiology 11:1878-1896.
• KAYTOUE-UBERALL M, DUPLESSIS S, KUZNETSOV SO, NAPOLI A (2009) Two FCA-based methods for mining gene expression data. In Proceedings of the 7th International Conference on Formal Concept Analysis (ICFCA 2009), Ferre S & Rudolph S (eds), published in Lecture Notes in Artificial Intelligence, Vol 5548, Springer, Berlin, Germany. Pp-251-266. DOI: 10.1007/978-3-642-01815-2_19
• ROUXEL T, DUPLESSIS S, VIAUD M, AMSELEM J, LEBRUN M-H (2009) La génomique fongique: la génomique des champignons pathogènes des plantes en France. Biofutur 296:31-35.
2008
 • KOHLER A, RINALDI C, DUPLESSIS S, BAUCHER M, GEELEN D, HELLSTEN U, DUCHAUSSOY F, DIFAZIO S, BOERJAN W, MARTIN F (2008) Genome-Wide identification of NBS resistance genes in Populus trichocarpa. Plant Molecular Biology 66:619-636. Cover of PMB april 2008 issue.
• KOHLER A, RINALDI C, DUPLESSIS S, BAUCHER M, GEELEN D, HELLSTEN U, DUCHAUSSOY F, DIFAZIO S, BOERJAN W, MARTIN F (2008) Genome-Wide identification of NBS resistance genes in Populus trichocarpa. Plant Molecular Biology 66:619-636. Cover of PMB april 2008 issue.
• KEMPPAINEN M, DUPLESSIS S, MARTIN F, PARDO AG (2008) T-DNA insertion, plasmid rescue and integration analysis in the model mycorrhizal fungus Laccaria bicolor. Microbial Biotechnology 1:258-269.
• MARTIN F, AERTS A, AHREN D, BRUN A, DUCCHAUSSOY F, KOHLER A, LINDQUIST E, SALAMOV A, SHAPIRO HJ, WUYTS J, BLAUDEZ D,BUEE M, BROKSTEIN P, CANBACK B, COHEN D, COURTY PE, COUTINHO PM, DANCHIN EGJ, DELARUELLE C, DETTER JC, DEVEAU A, DIFAZIO S, DUPLESSIS S, FRAISSINET L, LUCIC E, FREY-KLETT P, FOURREY C, FEUSSNER I, GAY G, GIBON J, GRIMWOOD J, HOEGGER P, JAIN P, KILARU S, LABBE J, LIN YC, LE TACON F, MARMEISSE E, MELAYAH D, MONTANINI B, MURATET M, NEHLS U, NICULITA H, OUDOT LE-SECQ MP, PEREDA V, PETER M, QUESNEVILLE H, RAJASHEKAR B, REICH M, ROUHIER N, SCHMUTZ J, YIN T, CHALOT M, HENRISSAT B, KUES U, VAN DE PEER Y, PODILA G, POLLE A, PUKKILA P, RICHARDSON PM, ROUZE P, SANDERS I, STAJICH J, TUNLID A, TUSKAN G, GRIGORIEV IV (2008) Symbiosis insights from the genome of the mycorrhizal basidiomycete Laccaria bicolor. Nature 452:88-92.
• KAYTOUE-UBERALL M, DUPLESSIS S, NAPOLI A (2008) Using formal concept analysis for the extraction of groups of co-expressed genes. In Proceedings of the 2nd International Conference on Modelling, Computation and Optimization in Information Systems and Management Sciences (MCO’08), Thi HAL, Bouvry P & Dinh TP (eds), published in Communications in Computer and Information Science, Springer, Berlin, Germany. Pp-439-449. DOI: 10.1007/978-3-540-87477-5_47
• DUPLESSIS S, KUHN H (2008) Meeting report: Secretomic climax in plant-fungal interactions. The New Phytologist 179:907-910. Corresponding author.
• KAYTOUE-UBERALL M, DUPLESSIS S, NAPOLI A (2008) Toward the discovery of itemsets with significant variations in gene expression matrices. Proceedings of the 1st joint meeting of the Société Francophone de Classification and the Classification and Data Analysis Group of SIS (SFC-CLADAG), June 11-13th 2008, Caserta, Italy. pp333-336.
2007
• RINALDI C, KOHLER A, FREY P, DUCHAUSSOY F, NINGRE N, COULOUX A, WINCKER P, LE THIEC D, FLUCH S, MARTIN F, DUPLESSIS S (2007) Transcript Profiling of Poplar Leaves upon Infection with Compatible and Incompatible Strains of the Foliar Rust Melampsora larici-populina. Plant Physiology, 144:347-366. Corresponding author.
• MARTIN F, KOHLER A, DUPLESSIS S (2007) Living in harmony in the wood underground: ectomycorrhizal genomics. Current Opinion in Plant Biology 10:204-210. Review.
2006
 • Tuskan G, DiFazio S, Bohlmann J, Grigoriev I, Hellsten U, Jansson S, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen G-L, Cooper D, Coutinho P, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Déjardin A, dePamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y, Henrissat B, Holligan D, Holt R, Huang W, Islam-Faridi N, Jones S, Jones-Rhoades M, Jorgensen R, Joshi C, Kangasjärvi J, Karlsson J, Kelleher C, Kirkpatrick R, Kirst M, Kohler A, Kalluri U, Larimer F, Leebens-Mack J, Leplé J-C, Locascio P, Luo Y, Lucas S, Martin F, Montanini B, Napoli C, Nelson DR, Nelson C, Nieminen K, Nilsson O, Peter G, Philippe R, Pilate G, Poliakov A, Razumovskaya J, Richardson P, Rinaldi C, Ritland K, Rouzé P, Ryaboy D, Schmutz J, Schrader J, Segerman B, Shin H, Siddiqui A, Sterky F, Terry A, Tsai C, Uberbacher E, Unneberg P, Vahala J, Wall K, Wessler S, Yang G, Yin T, Douglas C, Sandberg G, Van de Peer Y, Rokhsar D. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray ex Brayshaw). Science 313:1596-1604.
• Tuskan G, DiFazio S, Bohlmann J, Grigoriev I, Hellsten U, Jansson S, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen G-L, Cooper D, Coutinho P, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Déjardin A, dePamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y, Henrissat B, Holligan D, Holt R, Huang W, Islam-Faridi N, Jones S, Jones-Rhoades M, Jorgensen R, Joshi C, Kangasjärvi J, Karlsson J, Kelleher C, Kirkpatrick R, Kirst M, Kohler A, Kalluri U, Larimer F, Leebens-Mack J, Leplé J-C, Locascio P, Luo Y, Lucas S, Martin F, Montanini B, Napoli C, Nelson DR, Nelson C, Nieminen K, Nilsson O, Peter G, Philippe R, Pilate G, Poliakov A, Razumovskaya J, Richardson P, Rinaldi C, Ritland K, Rouzé P, Ryaboy D, Schmutz J, Schrader J, Segerman B, Shin H, Siddiqui A, Sterky F, Terry A, Tsai C, Uberbacher E, Unneberg P, Vahala J, Wall K, Wessler S, Yang G, Yin T, Douglas C, Sandberg G, Van de Peer Y, Rokhsar D. (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray ex Brayshaw). Science 313:1596-1604.
• SELOSSE M-A, DUPLESSIS S (2006) Meeting report: More complexity in the mycorrhizal word. The New Phytologist172:600-604.
• KOHLER A, PETER M, DUPLESSIS S, MARTIN F (2006) Microbial growth and visualisation of bacteria and fungi: Synthesis of ectomycorrhiza. In Hanbook of methods used in Rhizosphere Research, Luster J, Finlay RD (eds), Swiss Federal Research Institute WSL, Birmendorf, Suisse. pp399-400.
• PETER M, KOHLER A, DUPLESSIS S, MARTIN F (2006) Identification of single species and communities: RNA extraction from ectomycorrhizas. In Hanbook of methods used in Rhizosphere Research, Luster J, Finlay RD (eds), Swiss Federal Research Institute WSL, Birmendorf, Suisse. pp457-459.
• PETER M, KOHLER A, DUPLESSIS S, MARTIN F (2006) Gene expression of single species and communities: Large-scale analysis of gene expression in ectomycorrhizas. In Hanbook of methods used in Rhizosphere Research, Luster J, Finlay RD (eds), Swiss Federal Research Institute WSL, Birmendorf, Suisse. pp482-483.
2005
• GABELLA S, ABBÀ S, DUPLESSIS S, MONTANINI B, MARTIN F, BONFANTE P (2005). Transcript Profiling Reveals Novel Marker Genes Involved in Fruiting Body Formation in Tuber borchii. Eukaryotic Cell 4:1599-1602.
• PARDO A G, KEMPPAINEN M, VALDEMOROS D, DUPLESSIS S, MARTIN F, TAGU D (2005). T-DNA transfer from Agrobacterium tumefaciens to the ectomycorrhizal fungus Pisolithus microcarpus. Revista Argentina de Microbiología 37:69-72.
• GUPTA P, DUPLESSIS S, WHITE H, KARNOSKY D F, MARTIN F, PODILA G (2005). Gene expression patterns of trembling aspen trees following long-term exposure to interacting elevated CO2 and tropospheric O3. The New Phytologist 167:129-142. co-1st author.
• DUPLESSIS S, COURTY P-E, TAGU D, MARTIN F (2005). Transcript patterns associated with ectomycorrhiza development in Eucalyptus globulus and Pisolithus microcarpus.The New Phytologist 165:599-611.
2004
• ROUHIER N, GELHAYE E, GUALBERTO J M, JORDY M-N, DE FAY E, HIRASAWA M, DUPLESSIS S, LEMAIRE S D, FREY P, MARTIN F, MANIERI W, KNAFF D B, JACQUOT J-P (2004). Poplar Peroxiredoxin Q. A Thioredoxin-Linked Chloroplast Antioxidant Functional in Pathogen Defense. Plant Physiology 134:1027-1038.
• MARTIN F, DUPLESSIS S, KOHLER A, TAGU D (2004) Chapter 4. Exploring the transcriptome of the ectomycorrhizal symbiosis. In: Molecular Genetics and Breeding of Forest Trees (Kumar S, Fladung M, edts), Haworth’s Food Products Press, New York, pp. 81-109.
• KOHLER A, PETER M, JAMBOIS A, COURTY PE, DUPLESSIS S, LAPEYRIE F, MARTIN F (2004) Exploring the transcriptome of the ectomycorrhizal symbiosis. Meeting abstract APS 2004, Phytopathology 94:S133.
2003
• PETER M, COURTY P-E, KOHLER A, DELARUELLE C, MARTIN D, TAGU D, FREY-KLETT P,DUPLESSIS S, CHALOT M, PODILA G, MARTIN F (2003). Analysis of expressed sequence tags from the ectomycorrhizal basidiomycetes Laccaria bicolor and Pisolithus microcarpus. The New Phytologist 159:117-129.
• HUGOT K & DUPLESSIS S (2003) Réseaux d’ADN : puces à ADN, filtres d’ADNc. In Principes des Techniques de Biologie Moléculaire, 2nde édition revue et augmentée (D. Tagu & C. Moussard, éds), INRA Editions. pp 65-75.
2002
• LACOURT I, DUPLESSIS S, ABBA S, BONFANTE P, MARTIN F (2002). Isolation and characterization of differentially expressed genes in the mycelium and fruit body of Tuber borchii. Applied and Environmental Microbiology 68:4574-4582. co-1st author.
• BARKER SJ, DUPLESSIS S, TAGU D (2002). The application of genetic approaches for investigations of mycorrhizal symbioses. Plant and Soil 244:85-95.
• DUPLESSIS S, TAGU D, MARTIN F (2002). Living together underground : a molecular glimpse of the ectomycorrhizal symbiosis In Molecular biology of fungal development (Osiewacz, ed.). Mycology Series, Dekker & Dekker, New York, pp 297-323.
• MARTIN F, DUPLESSIS S, DITENGOU F A, LAGRANGE H, VOIBLET C, LAPEYRIE F (2002). Rhizospheric signals and early molecular events in the ectomycorrhizal symbiosis InPlant signal transduction : frontiers in molecular biology(Scheel D & Wasternanck C, edts). Oxford University Press, pp 272-288.
2001
• MARTIN F, DUPLESSIS S, DITENGOU F, LAGRANGE H, VOIBLET C, LAPEYRIE F (2001). Developmental cross talking in the ectomycorrhizal symbiosis : signals and communication genes. New Phytologist151:145-154.
• DUPLESSIS S, SORIN C, VOIBLET C, PALIN B, MARTIN F, TAGU D (2001). Cloning and expression analysis of a new hydrophobin cDNA from the ectomycorrhizal basidiomycete Pisolithus. Current Genetics39:335-339.
• VOIBLET C, DUPLESSIS S, ENCELOT N, MARTIN F (2001). Identification of symbiosis-regulated genes in Eucalyptus globulus-Pisolithus tinctorius ectomycorrhiza by differential hybridization of arrayed cDNAs. The Plant Journal 25:181-192. co-1st author.
• DUPLESSIS S (2001) PhD Thesis (in french) – Caractérisation par ingénierie génomique des profils d’expression génique de Pisolithus tinctorius et d’Eucalyptus globulus au cours du développement de la symbiose ectomycorhizienne. Thèse de Doctorat, Université Henri Poincaré-NancyI. pp105.
• DUPLESSIS S (1998) MSc (in french) – Caractérisation et étude de l’expression de gènes des voies de transduction chez le champignon ectomycorhizien Pisolithus tinctorius. Diplôme d’Etudes Approfondies, Université Henri Poincaré-Nancy I. pp20.
• DUPLESSIS S (1997) MSc (in french) Etude des composés phénoliques de la symbiose Betula pendula–Paxillus involutus par HPLC au cours des stades précoces de mycorhization. Mémoire de Maîtrise de Biologie Cellulaire et Physiologie Végétale, Université Henri Poincaré-Nancy I. pp25.
CONFERENCES & MEETINGS
• DUPLESSIS S, PETRE B, GERMAIN H, HECKER A, FREY P, HALKETT F, DE MITA S, JOLY DL, HACQUARD S, SEGUIN A, ROUHIER N. Investigating virulence effectors in the poplar-poplar rust pathosystem. 30th New Phytologist Symposium Immunomodulation by plant-associated organisms, Fallen Leaf Lake, CA, USA, September 2012 (Invited Speaker)
• DUPLESSIS S. Genomics of the poplar-poplar rust interaction: an emerging model pathosystem in forest pathology. 1st Symposium of the Annals of Forest Science journal, Champenoux, France, March 2012 (Invited Speaker)
• DUPLESSIS S. Candidate effectors of Melampsora larici-populina, the causal agent of the poplar leaf rust disease. REID network, Fungal Pathogens group meeting, Paris, France, december 2011 (speaker).
• DUPLESSIS S. Candidate effectors of Melampsora larici-populina, the causal agent of the poplar leaf rust disease. Effectome network 4th meeting, Lauret, France, september 2011 (speaker).
• DUPLESSIS S. Comparative genomic analysis of rust fungi. Basidiomycete meeting, satellite workshop of the 2011 JGI User Meeting, Walnut Creek, CA, USA, march 2011 (Speaker).
• DUPLESSIS S, HACQUARD S, JOLY DL, LIN Y-C, AND THE MELAMPSORA GENOME CONSORTIUM. Genome-wide analysis of the secretome repertoire in the poplar rust pathogen Melampsora larici-populina. 26th Fungal Genetics Conference, Asilomar, CA, USA, march 2011 (Poster).
• DUPLESSIS S. Les interactions Arbres-Champignons à l’ère de la génomique : Emergence du pathosystème modèle Peuplier/Melampsora. Journée scientifique de l’IFR110 EFABA, Nancy, France, December 2010 (Speaker).
• DUPLESSIS S. Candidate rust effectors in the secretome of the obligate fungal biotroph Melampsora larici-populina. Effectome Network 3rd Meeting, Lauret, France, October 2010 (Speaker).
• DUPLESSIS S. The poplar-rust interaction through the lens of genomics: an emerging model pathosystem in forest pathology. Laurentian Forestry Centre, Canadian Forest Service, Québec, Canada, May 2010 (seminar on invitation).
• DUPLESSIS S. Chasing Effectors in the Genomes of Fungal Symbionts and Pathogens of Trees. 110th General Meeting of the American Society for Microbiology. Division X Lectures, San Diego, CA, USA, May 2010 (invited speaker).
• Duplessis S, Lin Y-C, Tisserant E, Hilselberger B, Hacquard S, Joly D, Veneault-Fourrey C, Feau N, Tangay P, Kohler A, Lindquist E, Cantarel B, Couthinho P, Henrissat B, Frey P, Murat C, Amselem J, JGI Sequencing and Assembling Teams, Aerts A, Rouzé P, Van de Peer Y, Hamelin R, Grigoriev I, Martin F. The genome sequence of the poplar rust pathogen Melampsora larici-populina. 4th IUFRO International meeting on Rusts of Forest Trees, Florence, Italy, May 2010 (invited speaker). Abstract published in Phytopathol Mediterr 49:421-434.
• Hacquard S, Delaruelle C, Tisserant E, Legué V, Kohler A, Frey P, Martin F, Duplessis S. Laser capture microdissection of uredinia formed by Melampsora larici-populina revealed a transcriptional switch between biotrophy and sporulation. 4th IUFRO International meeting on Rusts of Forest Trees, Florence, Italy, May 2010 (Poster). Abstract published in Phytopathol Mediterr 49:421-434.
• Fernandez D, Tisserant E, Talhinhas P, Azinheira H, Petitot A-S, Duplessis S. Identification of Hemileia vastatrix in planta expressed genes through 454-pyrosequencing of rust-infected Coffea arabica leaf tissues. 4th IUFRO International meeting on Rusts of Forest Trees, Florence, Italy, May 2010 (Poster). Abstract published in Phytopathol Mediterr 49:421-434.
• DUPLESSIS S. Chasing effectors in the genomes of fungal symbionts and pathogens of trees. European Conference on Fungal Genetics, ECFG10, Netherlands, April 2010 (invited speaker).
• DUPLESSIS S. The genome sequence of the poplar leaf rust Melampsora larici-populina. Basidiomycete meeting, mini-symposium at the JGI User Meeting. Walnut Creek, CA, USA, March 2010 (Speaker).
• DUPLESSIS S. The genome sequence of the poplar leaf rust Melampsora larici-populina. 8èmes rencontres de Phytopathologie-Mycologie de la Société Française de Phytopathologie, JJC2010. Aussois, January 2010 (Speaker).
• DUPLESSIS S. The poplar-rust interaction through the lens of genomics: an emerging model pathosystem in forest pathology. University of Helsinki, December 2009 (seminar on invitation)
• DUPLESSIS S. The poplar-rust interaction through the lens of genomics: an emerging model pathosystem in forest pathology. 3rd UPRA Meeting, Nancy, France, November 2009 (invited speaker)
• DUPLESSIS S, HACQUARD S, JOLY D, FEAU N, LIN Y-C, TISSERANT E, FREY P, HAMELIN R, MARTIN F, JGI SEQUENCING & ASSEMBLING STAFF. Chasing effectors in the secretome of Melampsora larici-populina, the causal agent of the poplar leaf rust. 22nd New Phytologist Symposium, Versailles, France, September 2009 (invited speaker)
• DUPLESSIS S. Analyse moléculaire du sécrétome de Melampsora larici-populina, agent de la rouille foliaire du peuplier, et de Laccaria bicolor, symbiote ectomycrorhizien du peuplier. 7ème colloque national de la Société Française de Phytopathologie. Lyon, France, June 2009 (invited speaker of session ‘thèmes émergents en phytopathologie et interactions plantes-microorganismes)
• DUPLESSIS S. Génomique des interactions arbres-champignons biotrophes : recherche de marqueurs moléculaires des résistance qualitative et quantitative à Melampsora larici-populina chez le peuplier par des approches globales. IPMSV, INRA Antibes, France, May 2008 (seminar on invitation)
• DUPLESSIS S. The genome sequence of Melampsora larici-populina, the causal agent of the poplar rust disease. Basidio3 workshop on basidiomycete whole-genome sequences, Edinburgh, UK, april 2008 (Speaker).
• DUPLESSIS S, HACQUARD S, TICE H, DALIN H, LUCAS S, SHAPIRO H, PANGILINAN J, SCHMUTZ J, OUDOT-LE SECQ M-P, FREY P, RICHARDSON P, MARTIN F, Genome-wide analysis of secreted proteins encoding genes in Melampsora larici-populina, the causal agent of the poplar foliar rust disease. ECFG9, 9th European Conference on Fungal Genetics, Edinburgh, UK, april 2008 (poster)
• DUPLESSIS S, RINALDI C, KOHLER A, FREY P, LALANNE C, JORGE V, DOWKIW A, GUÉRIN V, FAIVRE-RAMPANT P, BRESSON A, PAOLUCCI I, PLOMION C, BASTIEN C, MARTIN F. Recherche de marqueurs moléculaires des résistances qualitative et quantitative à Melampsora larici-populina chez le peuplier par des approches globales. Journées Jean Chevaugeon. 7ème Rencontres de Phytopathologie/Mycologie de la SFP, Aussois, France, January 2008 (invited speaker)
• DUPLESSIS S. The sequencing of the Populus community genome for a better understanding of the tree-microbe interactions. Talca University, Chile, November 2007 (seminar on invitation).
• DUPLESSIS S. The sequencing of the Populus community genome for a better understanding of the tree-microbe interactions. 2nd UPRA Meeting, Umea, Sweden, October 2007 (invited speaker).
• DUPLESSIS S. Genomics of Tree-Microbe Interactions: genome sequencing, annotation & transcriptomics. Journées de bioinformatique du LORIA, Nancy, France, France, September 2007 (Speaker).
• DUPLESSIS S Genomics of the interaction between Populus and the poplar leaf rust, Melampsora larici-populina. Uppsala University, Sweden, April 2007 (seminar on invitation).
• DUPLESSIS S, LACCARIA GENOME CONSORTIUM, JOINT GENOME INSTITUTE. Annotation of signaling pathways genes families in the ectomycorrhizal basidiomycete Laccaria bicolor reveals an expansion of a G-proteins encoding genes family. Fungal Genetics Conferences, Asilomar, CA, USA, March 2007 (speaker).
• DUPLESSIS S, JAIN P, GROSDEMANDE A, DUCHAUSSOY F, MURATET M, PODILA G, MARTIN F. Annotation of signalling pathways genes families in the ectomycorrhizal basidiomycete Laccaria bicolorreveals an expansion of a G-proteins encoding genes family. IXth International Fungal Biology Conference & 16th New Phytologist Symposium, Nancy, France, September 2006 (poster)
• KOHLER A, PEREDA V, DUPLESSIS S, MARTIN F. Transcript patterns associated with ectomycorrhiza formation in poplar. 5th International Conference On Mycorrhiza, Granada, Spain, July 2006 (invited speaker).
• DUPLESSIS S, KOHLER A, RINALDI C, FREY P, MARTIN F. Dissecting the early molecular events in poplar leaves upon interaction with rust. XL annual meeting of the Argentine Society for Biochemistry and Molecular Biology. Iguazu, Missiones, Argentina, December 2004 (Poster).
• DUPLESSIS S, KOHLER A, RINALDI C, FREY P, MARTIN F. Characterization of the early response of poplar to rust infection using expression profiling. 12th New Phytologist Symposium on Functional Genomics of Environmental Adaptation in Populus. Gatlinburg TE, USA, October 2004 (invited speaker).
• RINALDI C, KOHLER A, DUPLESSIS S, FREY P, MARTIN F. Isolation and expression of a host- response gene family encoding thaumatin-like proteins in incompatible poplar-rust fungus interaction. 12th New Phytologist Symposium on Functional Genomics of Environmental Adaptation in Populus. Gatlinburg TE, USA, October 2004 (Poster).
• DUPLESSIS S. L’apport de la bioinformatique pour l’étude des interactions entre les arbres et les micro-organismes. 2ème journée de conférence de l’association DGIN, Université Henri Poincaré, Nancy, France, September 2004 (speaker).
• DUPLESSIS S. Exploring the transcriptome of the ectomycorrhizal symbiosis. Gordon Research Conferences on Cellular and Molecular Fungal Biology. Plymouth NH, USA, June 2004 (invited speaker).
• DUPLESSIS S. Transcriptome studies of tree/fungi interactions: transcriptome analysis of symbiotic and pathogenic interactions in forest trees. Uppsala University, Sweden, September 2003 (seminar on invitation).
• DUPLESSIS S, KOHLER A, ROUHIER N, FREY P, JACQUOT J-P, MARTIN F. Gene expression profiling in poplar leaves upon infection with rust. IUFRO Tree Biotechnology Meeting, Umea, Sweden, June 2003 (invited speaker).
• DUPLESSIS S, COURTY P-E, TAGU D, MARTIN F. Analyse du transcriptome de Pisolithus microcarpuset d’Eucalyptus globulus au cours du développement de la symbiose ectomycorhizienne. Séminaire annuel du Groupe d’Etude de l’Arbre ‘Réaction de l’arbre aux symbioses, parasites et ravageurs’, Nancy, France, May 2003 (speaker).
• DUPLESSIS S. Caractérisation des profils d’expression génique de Pisolithus microcarpus et d’Eucalyptus globulus au cours du développement de la symbiose ectomycorhizienne par une approche de génomique fonctionnelle : principes et limites des techniques mises en jeu, UPRES-EA 3569, Université de Lille, France, April 2003 (seminar on invitation).
• DUPLESSIS S, COURTY P-E, TAGU D, MARTIN F. Analyse du transcriptome de Pisolithus microcarpuset d’Eucalyptus globulus au cours du développement de la symbiose ectomycorhizienne. Journées du réseau de mycologie, Société Française de Microbiologie, Nancy, France, January 2003(speaker).
• DUPLESSIS S, COURTY P-E, TAGU D, MARTIN F. Transcriptome profiling indicates that multiple metabolic pathways are activated during the development of the Eucalyptus/Pisolithus ectomycorrhiza. 10th New Phytologist Symposium, Functional Genomics of Plant-Microbe Interactions, Nancy, France, October 2002 (invited speaker).
• DUPLESSIS S, ROUHIER N, FREY P, KOHLER A, JACQUOT J-P, MARTIN F. Analyse du transcriptome du pathosystème Populus x interamericana / Melampsora larici-populina. JJC, IVème Rencontres de Mycologie-Phytopathologie, Aussois, France, March 2002 (speaker).
• DUPLESSIS S, MARTIN F. Monitoring Eucalyptus globulus and Pisolithus tinctorius gene expression during the ectomycorrhizal symbiosis development using cDNA arrays and cluster analysis. Wood, Breeding, Biotechnology and industrial expectation, Bordeaux, France, June 2001 (poster).
• DUPLESSIS S. Caractérisation par ingénierie génomique des profils d’expression génique de Pisolithus tinctorius et d’Eucalyptus globulus au cours du développement de la symbiose ectomycorhizienne. Laboratoire de génétique, Pierroteon, May 2001 (seminar on invitation).
• DUPLESSIS S, VOIBLET C, MARTIN F. Monitoring differential gene expression in Eucalyptus globulus-Pisolithus tinctorius ectomycorrhiza using cDNA arrays. Fifth European Conference on Fungal Genetics, Arcachon, France, April 2000 (poster).
• DUPLESSIS S, VOIBLET C, MARTIN F. Caractérisation et étude de l’expression de gènes des voies de transduction du champignon ectomycorhizien Pisolithus tinctorius. JJC, IIIème Rencontres de Mycologie-Phytopathologie, Aussois, France, March 2000 (speaker).
• DUPLESSIS S, VOIBLET C, TAGU D, MARTIN F. ESTs et Filtres d’ADNc : Des méthodologies permettant d’étudier le transcriptome de la symbiose ectomycorhizienne. 5ème Rencontres BMoléculaire des Ligneux, Montpellier, France, October 1999 (speaker).





